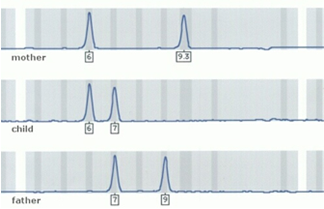
DNA profiling analyzes a number of specific polymorphic genetic markers, which are located on different chromosomes. These markers occur in pairs on our chromosomes and are transferred from parent to child, so that for each pair of markers, one is inherited from the mother and the other one from the father. This simple rule of inheritance is the basis for the DNA paternity test.
Specifically, DNA profiling through analysis of STR’s (Small Tandem Repeats) examines a set of polymorphic genetic markers-STR’s located on different chromosomes. Unlike older methods, involving analysis of VNTR’s, histocompatibility-HLA and blood group antigens, which are all now obsolete, genetic identification using specially developed STR markers is today the reference method (gold standard), both in investigation of paternity and in forensic applications.
Through the simultaneous analysis of several polymorphic markers-STR’s we achieve a ‘genetic fingerprint’ of each person that is practically unique in the world and identifies him completely. Thus, the comparison of such data through a special statistical analysis allows us to extract matching odds ratios, leading to the termine, and the exclusion of paternity.
Furthermore, the analysis of many polymorphic STR’s allows us also to establish paternity without the absolute need of a maternal sample and we can therefore establish paternity with a probability of >99.99% without testing the mother.
Note that the probability of paternity (W) cannot reach the absolute 100% (irrespective of the number of polymorphic STR markers analyzed), as the results are derived from a statistical calculation. Therefore, probability of paternity should not be confused with the probability of a laboratory error/accuracy of results.
For DNA paternity testing we use 16 internationally recommended and validated polymorphic STR markers (plus the non-polymorphic marker AMEL for gender determination), as well as other polymorphic STR markers of the X and Y chromosomes, where appropriate, thus achieving a very high probability (likelihood ratio). For the calculation of likelihood ratios we use a special software and the necessary STR frequency database in the Greek population, ensuring an outstanding reliability for our results.
For the DNA analysis we can use almost any type of biological sample/tissue in order to extract DNA from nucleated cells. The most frequently used sample is peripheral blood, particularly in adults, but we can also use equally well DNA derived from oral mucosal cells (buccal swab) or from root/hair follicle cells.